What drives dispersal for agricultural pests?
The larval form of click beetles, called “wireworms”, are agricultural pests for many important crops such as wheat, corn, and potatoes. Understanding population structure of these species can help us develop effective pest control methods. We investigated the population genetic structure of two wireworm species across the northwest US and southwest Canada, and found substantial population structure for each species, with some populations so divergent they might be considered separate species. The geographic distribution of populations likely results from reproductive isolation in Pleistocene glacial refugia, followed by range expansions after glaciers retreated. Many of the wireworm populations are separated by mountain ranges, which likely serve as modern-day barriers to gene flow. However, some populations are separated by no obvious modern-day or ancient barriers to gene flow, raising questions as to why those populations became distinct.
To read more about this work, check out our publication:
Does social structure drive genetic structure in Hawaiian spinner dolphins?
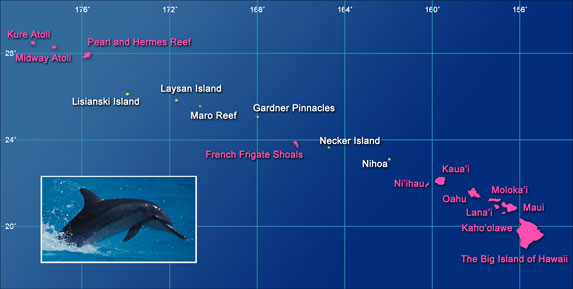
Map of the Hawaiian Archipelago with locations highlighted in pink where spinner dolphins regularly occur.
Social structure varies across the Hawaiian Archipelago for spinner dolphins: they have small populations (~100-300 dolphins) and high group stability in the far-northwestern Hawaiian Islands (Kure, Midway, and Pearl & Hermes), but they have larger population (~600 dolphins) and more fluid groups at the Big Island of Hawai‘i. We investigated the population genetic structure of spinner dolphins across the Hawaiian Archipelago, and, surprisingly, found evidence for high gene flow between the islands with stable groups (the far-northwestern islands), but limited gene flow to or from the island with the fluid social groups (Big Island ). High dispersal between the islands with strong group stability may be driven by evolutionary pressures to avoid inbreeding in very small populations.
To read more about this work, check out our publication:
Do ocean currents drive population structure in deepwater fish?
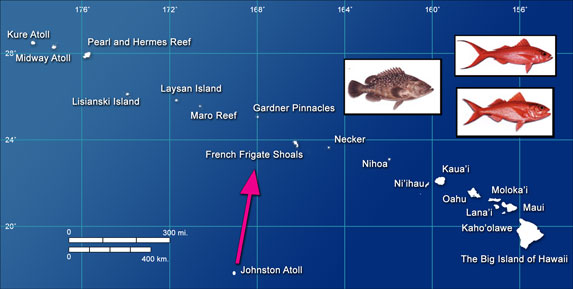
Map of the Hawaiian Archipelago and Johnston Atoll. The arrow illustrates larval dispersal from Johnston Atoll to the mid-archipelago.
Ocean currents are likely to play a major role in shaping the population genetic structure of fish by dispersing their larvae. To test this hypothesis for deepwater fish, we conducted population genetic analyses and oceanographic modeling of larval dispersal for three deepwater fish species across the Hawaiian Archipelago (“Ehu” Etelis marshi, “Onaga” Etelis coruscans, “Hawaiian grouper” Hyporthodus quernus). Genetic analyses indicated overall high connectivity across the Hawaiian Archipelago for all three fish species. However, a region in the middle of the archipelago (Gardner to Necker) had genetically distinct populations and/or high genetic diversity for all three fish. Our analyses indicated this pattern is likely driven by larval transport on ocean currents from Johnston Atoll, which is the closest landmass to Hawai‘i (~800km south).
To read more about this work, check out our publications:
What ecological factors control gene flow in holoplankton?
We might expect extensive gene flow across wide geographic areas for holoplanktonic species, because they spend their entire lives in the plankton, where ocean currents can disperse individuals widely and prevent the formation of isolated groups. However, our population genetic analyses found distinct populations separated by equatorial waters for the holoplanktonic copepod H. longicornis. Equatorial waters have high overall productivity, and we suspect these high productivity waters are poor habitat for this species, so that most individuals that enter these waters die. In this way, equatorial waters act as a barrier to gene flow, resulting in the presence of distinct populations on either side of the equator across the globe.
To read more about this work, check out our publications:


